Introduction
Dengue, an Aedes aegypti and Aedes albopictus mosquito-borne illness, emerged as a
global health problem in the 1960s [1]. One
study on the prevalence of dengue indicates that about half of the global population
is in danger, with an annual estimate of 100–400 million infections [1]. Another study estimates that about
This paper focuses on an alternative method of identifying dengue infection through a digital pathology approach from microscopic PBS images. The PBS analysis is a diagnostically relevant tool for evaluating various hematological disorders [8]. The manual interpretation of PBS through a microscope remains the backbone of hematological diagnostics, even though it is error-prone and time-consuming owing to its invaluable nature [9]. Automation of PBS analysis is a very active field of research that has motivated many researchers [8]. Automation can assist hematologists in yielding accurate and quick results, especially when there are tremendous amounts of samples to analyze. The digitization of PBS images using a digital microscope or a whole slide scanner, combined with the application of Artificial Intelligence (AI) – based tools, makes automated PBS image analysis feasible, limiting human intervention [8], [9].
In recent years, DL, a subset of AI, has been extensively and successfully used for automating various tasks, including healthcare-related tasks [10]. In particular, CNNs have gained popularity and become the main methodology for medical image analysis [10]. Digital pathology is one of the medical imaging areas where extensive use of CNNs is observed [10]. Whole-slide imaging systems are applied to digitize hematopathology/histopathology slides to generate images of high resolution [10]. The digitized slides are processed by incorporating CNN architectures to perform different computer vision tasks, which include classification (e.g., disease recognition), object detection (e.g., cell counting), and segmentation (e.g., nuclei identification) [10]. The ability of CNNs to extract high-level features without human supervision enables them to automatically learn the most discriminative features directly from the image [9], [11]. In particular, CNNs benefit from eliminating the tedious feature engineering process [12]. However, training from scratch demands a significantly huge labeled dataset. The inadequacy of freely accessible labeled images is one of the biggest hindrances in training CNNs for analysis of medical images (including digital pathology images) [10], [12]. An efficient solution in such a scenario is to employ pre-trained CNNs [12].
A pre-trained CNN is previously trained on millions of images from a generic dataset (e.g., ImageNet dataset) for a specific problem and could be employed to fix a fresh problem using a TL plan [10], [12]. TL signifies fine-tuning the model to fix a fresh problem [12]. The Fully Connected (FC) layers with randomly initialized weights are added to the pre-trained base model and trained on the new task-specific dataset during fine-tuning [13]. However, the base model weights are frozen to prevent them from getting updated during training to avert overfitting [13]. Commonly used CNN architectures for the analysis of medical images include AlexNet, VGGNet, ResNet, GoogLeNet, DenseNet, XceptionNet, and SqueezeNet [12]. These CNNs are trained to identify images into 1000 subcategories [14]. The advantages of transferred pre-trained CNNs include better performance, training with limited data, eliminating training from scratch, and speeding up the training process [12].
Lack of explainability is one main limitation of DL models [15], [16]. The logic behind the predictions made by these models is not clearly understood. Despite the excellent performance of the DL models in various healthcare applications, the medical fraternity still does not fully embrace them due to their black-box nature [17], [18]. The XAI technique was introduced to enable the medical fraternity to appreciate the philosophy behind the model predictions [17]. XAI explains the workings of the model and draws the users’ attention to the regions of the image that highly influence the model predictions [17]. The GradCAM XAI technique was built on the original CAM invented by Zhou et al. in 2015 [17], [19], [20]. GradCAM was developed to suit CNN architectures and is, therefore, more popular among DL models [15], [17].
A. Review of Related Literature
In recent years, enormous CNN architectures with TL have been put forward to analyze medical images, including digital pathology image analysis [10]. The following are some recently published articles that adopted pre-trained CNN architectures with a TL strategy for classifying leukocytes from digital microscopic PBS images.
Aziz et al. adopted a DL-based method for leukocyte classification (Munich AML
Morphology dataset) from blood smear images. Leukocytes were segmented using
K-means clustering in the color space -
The unique and distinctive contribution introduced by this proposed work is an XAI-integrated, computationally efficient deep-learning approach for automatically detecting dengue fever from digital microscopic PBS images. PBS analysis is a gold standard for diagnosing various hematological disorders, including dengue fever. The literature review revealed very few published works utilizing similar methodologies on comparable datasets. Most of the published work associated with automatic dengue detection was centered on tabular datasets containing symptoms/vital signs/blood profile data [33], [34], [35], [36]. Hence, this proposed work can potentially fill this gap in the literature. Only a few articles have been published on the automatic diagnosis of dengue from PBS images [37], [38]. In one of our published works [37], we employed MobileNetV2 (a pre-trained CNN) for extracting features of the lymphocyte nucleus. Further, these deep features were administered to popular supervised classifiers to detect dengue-infected smears against the normal ones. A classification accuracy of 95.74% was obtained with the Support Vector Machine (SVM). This paper presents an explainable DL approach for dengue detection from 100x digital microscopic PBS images using pre-trained CNNs with TL. Here, an end-to-end system performs the classification process, bypassing segmentation and feature extraction/selection. To the best of our knowledge, there has been no prior publication on the use of this technique for dengue detection from PBS images.
Significant contributions of this article are listed as follows:
Novel end-to-end computationally efficient DL system for automatically detecting dengue from PBS images.
Integration of GradCAM XAI technique that provides confidence to the clinicians in the DL model’s predictions.
Utilizes an original hospital dataset (i.e., microscopic PBS images of dengue-infected and normal controls) collected systematically under ethical clearance.
Methodology
This paper proposes an explainable DL approach for detecting dengue fever from microscopic images of blood smears. State-of-the-art pre-trained CNNs are used to lower the expenses of model training from scratch. Heat maps are generated using the GradCAM XAI technique to highlight which areas on the image are concentrated in the predictions of the CNN model. The clear-cut details concerning the data, and the implementation of the TL and the XAI are described here.
A. Image Dataset
The dataset adopted is authentic hospital data accumulated from the Hematology
Lab, Kasturba Hospital, Manipal. The data is acquired under ethical clearance
(114/2020) granted by the Institutional Committee. Normal blood samples are
collected from the Department of Immunohematology, Kasturba Hospital, Manipal,
from blood bank donors visiting the department. Figure 1 shows the dataset preparation process. The dataset contains
888 PBS images (446 dengue-infected and 442 normal controls) garnered from 116
Leishman-stained blood smear thin glass slides (60 dengue-infected and 56 normal
controls). A high-quality technology brightfield microscopic imaging system -
Olympus DP25 digital microscope facilitated with a 5-megapixel high-precision
digital camera and DP2-BSW software is used to capture the digital images of
exceptional quality (
The emphasis is on the lymphocytes (one among the five types of leukocytes) throughout the work. Dengue alters the morphology of the lymphocytes. Studies show that this is an important indicator for dengue diagnosis [39]. PBS images of dengue-infected and normal control subjects are shown in Figure 2.
B. Implementation Details
TL is executed on three state-of-the-art CNNs – ResNet50, MobileNetV3Small, and MobileNetV3Large and is utilized to discriminate dengue-infected smears from normal. The pipeline for dengue detection from PBS using transferred pre-trained CNNs is displayed in Figure 3.
1) Pre-Trained CNNs—ResNet50, MobileNetV3Small, and MobileNetV3Large
ResNet50 is the most common type of residual network that was introduced in
2015 to address the problems of vanishing gradient and performance
degradation associated with deep CNNs [40], [41]. It has 107
layers (49 convolution layers + an FC layer) and approximately 26
million parameters [42]. ResNet50
architecture begins with a convolution layer and is followed by 16 stacked
building blocks (residual) and terminates with an FC layer [43], [44]. Each residual block is a bottleneck block
consisting of a stack of three convolution layers (
MobileNetV3, introduced in 2019, is the latest version of lightweight MobileNets [45], [46]. MobileNetV3 was built upon the MobileNetV1 and V2 structures. MobileNetV1 uses lightweight depth-wise convolutions to reduce the number of parameters [47]. MobileNetV2 was built upon MobileNetV1 with a new resource-efficient feature added: Inverted residual blocks with linear bottlenecking structure [47]. To make the structure more accurate and efficient, MobileNetV3 was introduced. Major improvements in MobileNetV3 include the Squeeze and Excitation - attention module added to the residual block and the use of hard-swish non-linearity instead of ReLU [47]. Moreover, platform-aware network architecture search and NetAdapt algorithm are engaged to optimize the architecture at the block and layer levels, respectively [47], [48]. Furthermore, MobileNetV3 includes small and large versions that operate on the same principle but vary in depth and trainable parameters [45], [49]. These networks have drastically lower parameter counts (approximately 2.9 million for small and approximately 5.4 million for large) and can be effectively implemented on resource-constrained devices [50].
2) Transfer Learning
The transfer learning plan accommodates the pre-trained CNNs to the dengue dataset. The pre-trained CNNs ResNet50, MobileNetV3Small, and MobileNetV3Large are base models. TL signifies fine-tuning the CNNs to resolve a new problem [12]. During fine-tuning, the trainable layers with randomly initialized weights are added on top of the pre-trained base model and trained on the new task-specific dataset [13]. Trainable layers include a flattened layer, a dense layer with 512 neurons and ReLU activation, a batch normalization layer, a dropout layer with rate=20%, a dense layer with 256 neurons and ReLU activation, a dropout layer with rate=20%. The output is fed to the final dense layer with two classes and Softmax activation to classify. These layers are added after the frozen base model, as illustrated in Figure 4.
The architecture of transferred pre-trained CNNs based on (a) ResNet50 (b) MobileNetV3Small (c) MobileNetV3Large.
3) Hyperparameters
The hyperparameters used to train the transferred pre-trained CNN architectures are tabulated in Table 1. These hyperparameters are fine-tuned to attain the optimum performance. Dropout is used after each dense layer to mitigate overfitting by randomly dropping out 20% of the neurons during training. The Learn Rate is initialized to 0.0001, and a scheduler is employed to dynamically decay the Learn Rate by a factor of 0.1 when the validation loss does not improve over four consecutive epochs. The size of the batch is fixed to 32 due to memory constraints, and the network is trained with maximum epochs set to 40. The early stopping mode is triggered to halt the training when the validation loss does not improve over six consecutive epochs, and the best weights are restored.
The randomly initialized weights of the trainable layers are upgraded during
the training of the models, depending on the optimization algorithm, to
lessen the loss function [51].
Here, the loss function is cross-entropy. Cross-entropy loss is represented
by Eq. (1)
![]() \begin{equation*} L_{CE}=-\sum \nolimits
_{i=1}^{n} {t_{i}log\left ({p_{i} }\right)} \tag{1}\end{equation*}
\begin{equation*} L_{CE}=-\sum \nolimits
_{i=1}^{n} {t_{i}log\left ({p_{i} }\right)} \tag{1}\end{equation*}
In Eq. (1),
n is the number of classes, ![]() \begin{equation*} w: =w- \alpha \partial
L/\partial w \tag{2}\end{equation*}
\begin{equation*} w: =w- \alpha \partial
L/\partial w \tag{2}\end{equation*}
In Eq. (2),
w stands for the weight,
The pseudo-code for the implementation of transfer learning for the proposed work.
All the CNN models are executed using Keras (with TensorFlow backend) on Kaggle Notebooks with Nvidia GPU P100 Accelerator (15.9GB RAM) conducted on a 64-bit Laptop with an Intel Core i5-7200U CPU (8GB RAM).
4) GradCAM Explanations
GradCAM is the most extensively employed XAI technique for medical image analysis [17], [53]. It is very often coupled with DL models such as CNNs, which are popular for image recognition [15], [17]. The last convolution layer of a CNN model contains the most discriminative features with detailed spatial information [15], [17]. GradCAM uses the gradients of the class score with respect to the feature maps at the final convolution layer to generate heat maps [15], [17]. These heatmaps are then superimposed on the images, which help the users to see the areas of the image that are most valuable for the model predictions. GradCAM follows three steps [15], [18], [54] to generate the heat maps, as shown below:
Step 1:
Calculate the class score gradient c,
y^{c} A^{k} \frac {\partial y^{c}}{\partial A_{i,j}^{k}} Step 2:
Global average pool the gradients to obtain the weights
\alpha _{k}^{c} \begin{equation*} \alpha _{k}^{c}=\frac {1}{N}\sum \nolimits _{i} \sum \nolimits _{j} \frac {\partial y^{c}}{\partial A_{i,j}^{k}} \tag{3}\end{equation*} View Source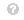 \begin{equation*} \alpha
_{k}^{c}=\frac {1}{N}\sum \nolimits _{i} \sum \nolimits
_{j} \frac {\partial y^{c}}{\partial A_{i,j}^{k}}
\tag{3}\end{equation*}
\begin{equation*} \alpha
_{k}^{c}=\frac {1}{N}\sum \nolimits _{i} \sum \nolimits
_{j} \frac {\partial y^{c}}{\partial A_{i,j}^{k}}
\tag{3}\end{equation*} In Eq. (3), N represents the pixels in the feature map.
Step 3:
The GradCAM map is then a weighted combination of the feature maps with an applied ReLU as given by Eq. (4).
\begin{equation*} M=\mathrm {RELU}\left ({\sum \nolimits _{k} \alpha _{k}^{c} A^{k} }\right) \tag{4}\end{equation*} View Source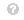 \begin{equation*} M=\mathrm
{RELU}\left ({\sum \nolimits _{k} \alpha _{k}^{c} A^{k}
}\right) \tag{4}\end{equation*}
\begin{equation*} M=\mathrm
{RELU}\left ({\sum \nolimits _{k} \alpha _{k}^{c} A^{k}
}\right) \tag{4}\end{equation*}
The ReLU activation preserves only the features that have a positive contribution to the class of interest.
Results and Discussion
All three models are assessed using the five-fold cross-validation scheme, wherein
the data is split randomly into training (80%) and validation (20%).
Further, the models are tested with unseen data. The test dataset contains 78 PBS
images, of which 37 are dengue-infected, and 41 are normal controls. Six popular
indices – Accuracy, Recall, Specificity, Precision, F1-score, and Area Under
the ROC Curve (AUC) are used to gauge the model’s performance. These
statistical metrics are derived from true positives (‘Dengue’
correctly classified; TP), true negatives (‘Normal’ correctly
classified; TN), false negatives (‘Dengue’ incorrectly classified as
‘Normal’; FN), and false positives (‘Normal’ incorrectly
classified as ‘Dengue’; FP) and are defined in Eq. (5)–(9).![]() \begin{align*} \mathrm {Accuracy}&=\frac {\mathrm
{TP+TN}}{\mathrm {TP+TN+FP+FN}} \times 100 \tag{5}\\ \mathrm
{Recall}&=\frac {\mathrm {TP}}{\mathrm {TP+FN}} \times 100 \tag{6}\\
\mathrm {Specificity}&=\frac {\mathrm {TN}}{\mathrm {FP+TN}} \times 100
\tag{7}\\ \mathrm {Precision}&=\frac {\mathrm {TP}}{\mathrm {FP+TP}}
\times 100 \tag{8}\\ \mathrm {F1 Score}&=\frac {\mathrm {2\ast Pre\ast
Sen}}{\mathrm {Pre+Sen}} \times 100 \tag{9}\end{align*}
\begin{align*} \mathrm {Accuracy}&=\frac {\mathrm
{TP+TN}}{\mathrm {TP+TN+FP+FN}} \times 100 \tag{5}\\ \mathrm
{Recall}&=\frac {\mathrm {TP}}{\mathrm {TP+FN}} \times 100 \tag{6}\\
\mathrm {Specificity}&=\frac {\mathrm {TN}}{\mathrm {FP+TN}} \times 100
\tag{7}\\ \mathrm {Precision}&=\frac {\mathrm {TP}}{\mathrm {FP+TP}}
\times 100 \tag{8}\\ \mathrm {F1 Score}&=\frac {\mathrm {2\ast Pre\ast
Sen}}{\mathrm {Pre+Sen}} \times 100 \tag{9}\end{align*}
Figure 6 shows the Accuracy and Loss plots of the Training and Validation for five folds of the transferred CNN based on MobileNetV3Small. Table 2 presents the classification accuracies of the transferred CNN models on the validation dataset for each of the five folds, along with the classification accuracy averaged over the five folds. Table 3 highlights the performance of the models during training and validation, averaging over five folds. Table 4 summarizes the detailed classification performance of the models on the validation dataset, averaging over five folds. Figure 7 reports the confusion matrices, and Table 5 reports the overall model’s performance on the test (unseen) data, averaging over the five folds.
Accuracy and Loss plots - Training (Blue) / Validation (Orange) for five folds of the transferred pre-trained CNN based on MobileNetV3Small.
Confusion matrices (Mean ± SD) for five-fold cross-validation on the test dataset of the transferred pre-trained CNN based on (a) ResNet50, (b) MobileNetV3Small, and (c) MobileNetV3Large.
While all models rendered good performance (above 98% overall classification accuracy), MobileNetV3Small is the recommended model for this classification problem due to its significantly less computationally demanding characteristics. MobileNetV3Small is characterized by limited parameters, i.e., approx. 2.9 million, and therefore is the least time-consuming and has the least resource requirement.
Figures 8 and 9 demonstrate the GradCAM localization of the areas of the image that are most valuable for the predictions. As a result, the proposed transferred pre-trained CNN models (for example, based on ResNet50) recognized the changes in the morphology of the lymphocytes in Figure 8 and highlighted it with an activation map, demonstrating that it is the noteworthy region of interest for the identification of Dengue class in the image. Instead, as indicated in Figure 9, the model for the Normal class did not highlight the lymphocyte region, indicating no morphological changes in the lymphocyte. The red/yellow color in the activation maps indicated more concentrated regions, and the less concentrated regions are with lighter colors approaching green/blue.
Activation map for Dengue class obtained from the GradCAM technique for the transferred pre-trained CNN based on ResNet50.
Activation map for Normal class obtained from the GradCAM technique for the transferred pre-trained CNN based on ResNet50.
The outcomes of this work are inspiring and demonstrate that the pre-trained CNNs have the capacity to yield commendable assistance in the PBS analysis for dengue diagnosis. This work is crucial and can contribute significantly to healthcare as this offers some level of explainability of the inner workings of the CNNs that clinicians may relate to. Moreover, this work is better in the context that it bypasses laborious steps like segmentation of lymphocyte nucleus, feature extraction, and feature ranking, in contrast to the classical machine learning methods. In one of our previous works [38], ten, morphological and GLCM features were extracted from the segmented lymphocyte nucleus. These features, when coupled with SVM, achieved the best classification with Accuracy, Recall, Specificity, Precision, F1 Score, and AUC of 93.62%, 92.59%, 95%, 96.15%, 94.34%, and 0.96, respectively. Furthermore, this work differs from works in that the authors have taken advantage of pre-trained CNNs as feature extractors and linked them to a separate machine-learning classifier. In another previous work [37], 1,000 deep and 177 Local Binary Pattern generated features were derived from the segmented lymphocyte nucleus. Deep features were derived by utilizing pre-trained CNN MobileNetV2 as a feature extractor. Feature selection algorithm ReliefF was used to select 100 important features. These features given to SVM yielded the best classification with accuracy, recall, specificity, precision, F1 Score, and AUC of 95.74%, 98.15%, 92.50%, 94.64%, 96.36%, and 0.98, respectively. The model’s performance can be further boosted by exploring different fine-tuning strategies.
There was a dearth of existing publications employing PBS images for dengue diagnosis, and the literature review revealed no published works from other research groups utilizing similar methodologies on comparable datasets. Most of the research work reported on automated diagnosis of dengue is by utilizing symptoms, vital signs, blood profile data, or a combination of these. Gambhir et al. proposed a PSO-optimized ANN for the diagnosis of dengue. With 16 attributes containing symptoms, vital signs, and blood profile data, authors classified the data and documented an accuracy, recall, and specificity of 87.27%, 68%, and 92.94%, respectively [55]. Mello-Roman et al. developed a symptom-based diagnostic model for dengue fever. With 38 attributes, including symptoms, the authors classified the data using MLP and documented an accuracy, recall, and specificity of 96%, 96%, and 97%, respectively [35]. Katta et al. used symptoms to develop an efficient model for dengue detection. The RF classifier yielded an accuracy and sensitivity of 94.39% and 95.60%, respectively [33]. Hoyos et al. developed a decision-support system for dengue diagnosis using the fuzzy cognitive map. With 22 features, including symptoms, vital signs, and blood profile data, the authors achieved a classification accuracy of 89.40% [36]. Table 6 summarizes the comparison of the performance of the proposed work with other works on automated detection of dengue published in the literature.
This work incorporates an end-to-end computationally efficient DL system for automatically detecting dengue from PBS. The system is integrated with GradCAM explainability and utilizes data sourced authentically from Kasturba Hospital, Manipal, under ethical clearance. Certainly, the entries in Table 6 depict that the proposed method for dengue diagnosis outperformed in comparison with the state-of-the-art studies published in the literature.
Conclusion and Future Work
The examination of the PBS is a powerful adjunct to other clinical procedures. In connection with dengue diagnosis, it can be a crucial add-on to the Complete Blood Count test and NS1 antigen capture and can substantially aid dengue diagnosis in low-resource settings. This work utilizes pre-trained CNNs for dengue fever detection from digital microscopic PBS images. The transfer learning strategy was successful in differentiating dengue-infected and normal smears. All three models rendered good performance with classification accuracy above 98%. Despite being less computationally expensive, the performance of the transferred pre-trained CNN based on MobileNetV3Small is at par with the other two models. Hence, transferred pre-trained CNN based on MobileNetV3Small is the preferred model for the proposed method of dengue diagnosis. Explainability is recognized as a key component for the acceptance of AI systems for clinical use. An explainability technique - GradCAM was integrated to the models to visualize the specific regions of the smears that were most dominant in making the predictive decisions. Promising results show that the developed models have the potential to provide high-quality support to haematologists by expertly executing tedious, repetitive, and time-consuming duties in hospitals and remote/low-resource settings. Future works could be focused on examining different fine-tuning strategies to facilitate performance improvement. The dataset size (multiple hospital data) could be further improved to increase the algorithm’s robustness. Moreover, a multiclass problem could be considered for analyzing dengue severity (normal/mild dengue/severe dengue). Furthermore, in the future, our method could be incorporated into mobile devices with a microscope attachment and utilized as a standalone product to screen for dengue in hospitals.
ACKNOWLEDGMENT
The authors thank the Department of Pathology, Kasturba Hospital, MAHE, Manipal, for providing the peripheral blood smear slides and a digital microscope to capture the peripheral blood smear images required for this work. They also would like to thank the Department of Immunohaematology, Kasturba Hospital, MAHE, Manipal, for providing blood samples from healthy blood bank donors.
















